
- +1 (714) 578-6100
Hours Mon - Fri, 07:00 AM - 06:00 PM (Pacific Time)
Since its advent in 1995, miniaturized microarray analysis has become a mainstay analytical technique of genomic, proteomic, pharmaceutical and clinical laboratories worldwide. The initial microarray chips, developed by Pat Brown at Stanford University, utilized oligonucleic acid probes deposited on a substrate to discover the genomic composition of cells. After this initial focus, the technology rapidly expanded to utilize other, related probes (such as DNA, RNA, peptides, carbohydrates).
In addition to expanding the types of molecular probes, the technology also increased the types of substrates to which probes can be attached. Glass slides, plastic/silica chips, and silica or polystyrene beads can now be used as the medium for constructing a microarray.

Microarray slides (Arrayit Corp).
Microarray Design and Experimentation
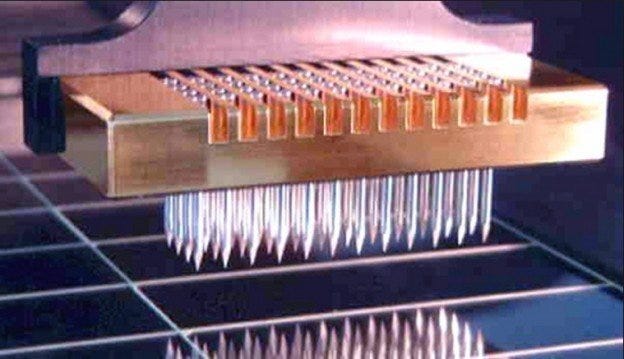
In general, a microarray consists of a group of micron-sized spots of a given probe “printed” in an ordered batch. These spots are single, known oligonucelotides, peptides or carbohydrates. They are immobilized on a surface, typically through hydrogen bonding. Once attached, a pool of proteins, nucleic acids or whole-cell lysates are labeled with a tag (fluorophore, metal, or chemiluminescent) and hybridized with the probes. After thorough washing, the degree of binding is optically examined.
This method measures the degree of hybridization, from which several conclusions can be made. Researchers utilize these data to determine genotypes, abundance of a given mRNA, protein presence and levels, and even small-molecule affinity.
There are myriad applications for microarrays. Microarray experimentation can generate large amounts of data in a short period of time. In fact, the rate-limiting step in microarray analysis has now become processing of the input. The “big data” generated through microarray analysis (as with other analytical techniques) are so extensive that the field of bioinformatics has concurrently evolved.
Lastly, relatively inexpensive equipment now exists for in-house printed microarrays, opening this method of investigation to almost any lab. Selecting the proper materials to be used for your microarray analysis should be carefully considered.
Selecting the Medium: Beads, Slides or Chips?
When selecting a medium for your microarray analysis, consider these factors. Laboratories have the option of purchasing pre-printed microarray chips and beads from manufacturers, or printing substrates in the lab. The pre-printed products are prepared with a known set of probes, designed specifically for a cell, tissue type or cellular process. Glass slides are typically used for in-house printing where users can select a unique set of probes for interrogation based on their needs.
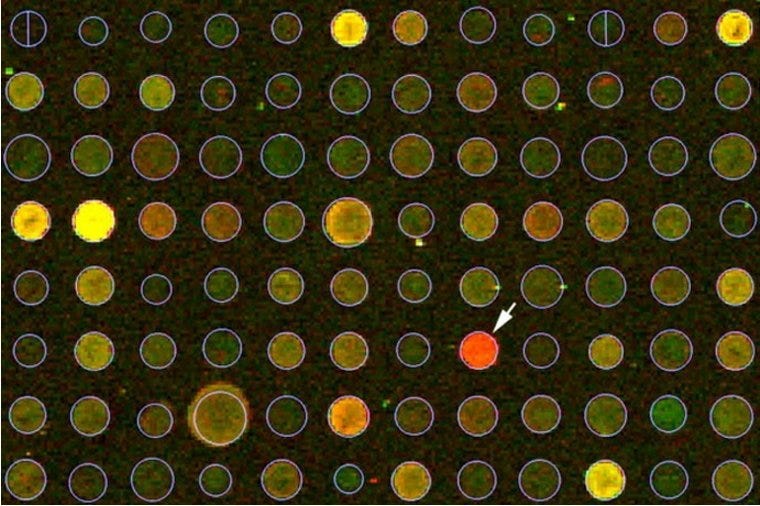
Microarray analysis (control = red).
Microarray slides are typically constructed of glass with an extremely low refractive index, manufactured within a cleanroom to prevent contamination. Application-specific surface chemistries coat the slides. Immobilization of probes is easily done using affordable instrumentation. The obvious benefit of in-house printing is the ability to select your own probes. Pre-printed microarrays will often contain probes that are not applicable to the study being performed by the researchers.
Due to the limited manufacturing capacity and type of technology used for in-house printing, these microarrays typically have lower hybridization efficiencies compared to those obtained from manufacturers. Labs that are processing large numbers of microarrays will benefit from the reduced cost of in-lab printing, while those performing low-quantity experimentation will likely find it more cost-effective to purchase pre-printed slides.
Pre-printed microarrays are typically spotted onto silica or plastic chips or immobilized onto beads. Plastic and silica chips have traditionally been used in the construction of microarrays. These chips benefit from a near-zero degree of reflection and intrinsic fluorescence, conditions that create background noise. In addition, these slides are manufactured in large quantities, and tightly controlled and tested for data quality and hybridization performance.

Microarray printing pin (Arrayit).
Bead-based assays also benefit from this degree of control. In addition, they can be conducted in solution, avoiding the need to handle fragile media with specialized hybridization and washing equipment. Having access to specialized multi-channel fluorescence instrumentation is the primary drawback of bead-based assays. Additionally, this assay technique can identify individual probes by comparing a ratio of two separate fluorophores, but measurements must be taken at a wavelength minimum. Despite these drawbacks, many labs will opt to purchase higher-efficiency pre-configured chips and beads that don’t require an in-house microarray printer.
Whether you choose to purchase microarray chips or beads or print your own microarrays, these experiments give you the opportunity to gather large amounts of data within a short period of time. Traditionally, proteomic and genetics laboratories were required to investigate genes and proteins one-at-a-time. The advent of microarrays has altered this dynamic so that now even the smallest laboratory can investigate an entire genome or proteome in a single experiment.
Terra Universal is the leading expert in the design and fabrication of ISO rated cleanrooms, furnishing and supplies.
Call (714) 459-0731
